The conversion of solar radiation to chemical energy by photosynthetic organisms provides the primary driving force for life on earth. Light energy is captured by a variety of pigments, usually bound to proteins, which vary with different types of organisms. We report here the 1.45 Angstrom resolution three-dimensional structure of one such pigment protein, C-phycocyanin, from Synechococcus elongatus, Kovrov strain.
This structure was the highest resolution achieved for any such phycobiliprotein (light-harvesting protein) at that time (2003). The level of resolution gained was made possible by implementing a novel crystallisation method whereby nucleation is decoupled from subsequent growth. This was done without touching the crystallisation drops throughout the crystallisation process.
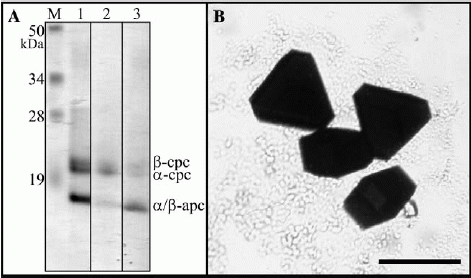
Fig. 1. (A) SDS–PAGE of the sample used for crystallisation trials (B) Examples of the large, well ordered, three-dimensional crystals used for Xray diffraction. Bar represents 250 um.
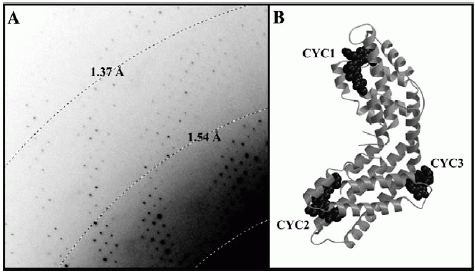
Fig. 2. (A) A typical diffraction pattern, showing extension beyond 1.37 Å.
(B) Ribbon representation of the S. elongatus C-PC heterodimer.
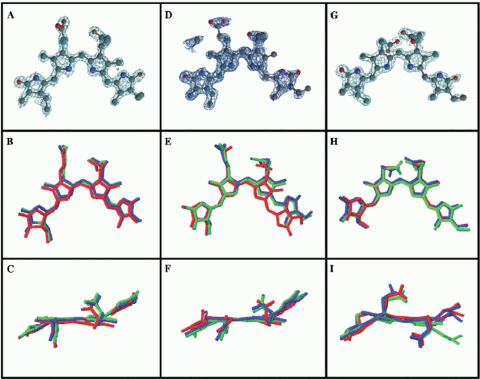
Fig. 3. The S. elongatus chromophore models CYC1 (panels A-C), CYC2 (D-F) and CYC3 (G-I). Top row: electron density around the chromophores. Middle: S. elongatus chromophores (blue) projected normal to the rings. Bottom row: viewed on edge. The lower 2 rows show the superimposition of chromophore models from F. diplosiphon (green) & C. caldarium (red).
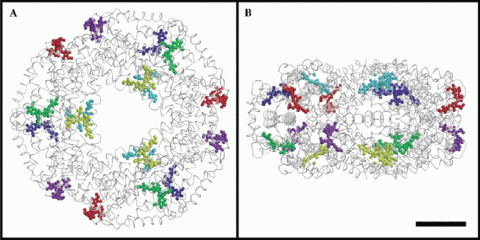
Fig. 4. In the phycobilisome and also in the crystal, the / subunits oligomerize to form a hexamer consisting of two ab3 trimers.

Diffraction patterns from C-PC crystals (e.g. Fig. 2A) identified the space group as R32 (No. 155) with unit-cell dimensions of a = b = 187.99 Å and c = 60.54 Å. This c-cell edge was very close to that of F. diplosiphon and C. caldarium, suggesting similar packing. Assuming a molecular weight of 35.6 kDa per / C-PC heterodimer, and one heterodimer per asymmetric unit, then Vm = 2.89 Å cubed/Da, implying a solvent content occupying ~ 42% of the total volume. Vm decreased to 2.81 Å cubed/Da on inclusion of the 3 covalently linked chromophores. Diffraction was observed beyond the 1.4 Å limit (Fig. 2A), with the data being reliable up to 1.45 Å. Relevant statistics are given in Table 1 above. Crystallographic data have been deposited with the RCSB structural database (http://www.rcsb.org) under codes 1JBO (capital ‘O’) for the co-ordinates and R1JBOSF for the structure factors.
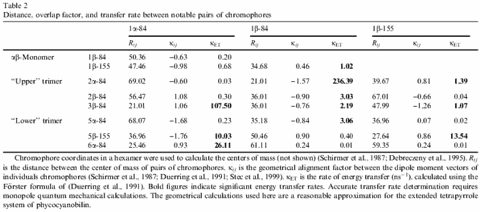
In the heterodimer, the separation and geometry of the three chromophores allows for only weak excitonic coupling. However, in the phycobilisome and also in the crystal, the subunits oligomerise to form a hexamer consisting of two ab3 trimers (see Fig.4). When 3 monomers are assembled into an ab3 trimer the local environments around CYC1 and CYC2 chromophores change considerably. In the monomer, the distance between these two chromophores is ~50 Å based on the centres of gravity of the conjugated portions of the chromophores. In the case of the trimer, the distance between the adjacent CYC1/CYC2 reduces to 21.0 Å, facilitating a stronger excitonic interaction between them (see Table 2 above). For inter-chromophore distances within the hexamer, energy transfer is also favoured between these chromophores to such an extent that we have calculated one of the highest transfer rates observed for such a protein, 236.4 per nanosecond. Furthermore, in the hexamer the CYC3 chromophores are close enough to permit efficient coupling, although this is less favourable compared to those within each trimer.
For this paper, Jon gratefully acknowledged he was in receipt of a Royal Society University Research Fellowship. Both he and the authors thanked the Biotechnology and Biological Research Sciences (BBSRC) for funding and the technical assistance of Karim Maghlaoui for preparation of cyanobacterial cultures as well as financial input from the Leverhulme Trust. We thanked the staff at the SRS (Daresbury) and Station 14.1 for their technical help, and to the UK Research Councils for the supply of beam-time.
© Journal of Structural Biology, 2003, Elsevier Science (USA). All rights reserved. doi:10.1016/S1047-8477(02)00609-3.